Figure 4
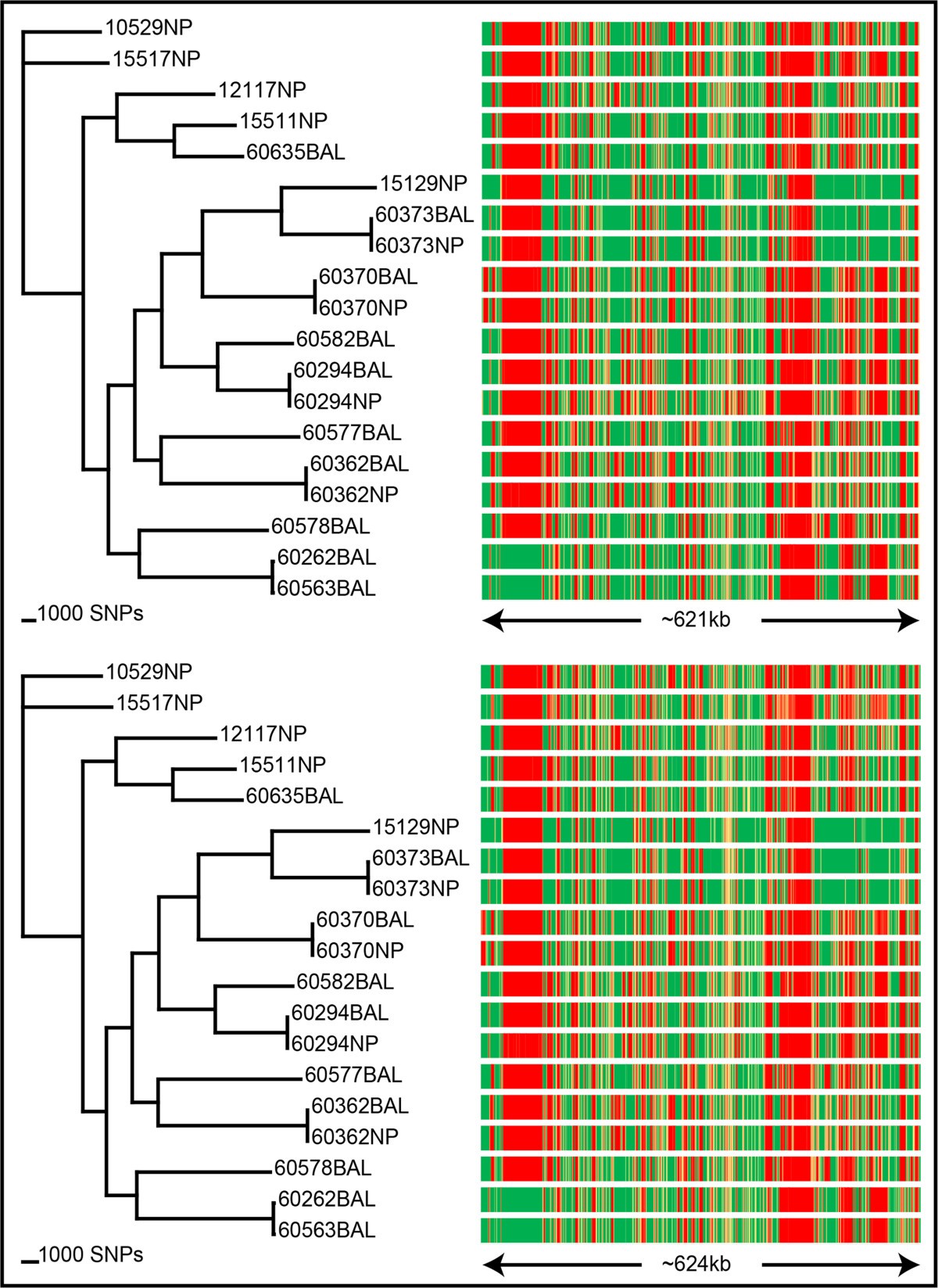
Comparison of Illumina and Ion PGM platforms using SPANDx. SPANDx was tested on 19 Australian Haemophilus influenzae strains[16] with both single-end Ion PGM and paired-end Illumina data. Strain 86-028NP[21] was used for reference alignment. From the Illumina data (top left), ~161,000 identified SNPs were used to construct a core genome SNP cladogram (CI = 0.47). From the Ion PGM data (bottom left), ~129,000 identified SNPs were used to construct a core genome SNP cladogram (CI = 0.48). The right-hand side panels show corresponding presence/absence data for each strain as described in Figure 3. For Illumina, 621 kb was found to be variable, compared with 624 kb with the Ion PGM data. Collectively, this comparison shows that SPANDx provides highly consistent haploid comparative genomic outputs across multiple NGS platforms.